Automatic Segmentation of 3D MRI Images of the Pituitary Gland for Medical Image Analysis
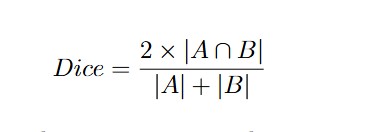

Project Title: Automatic Segmentation of 3D MRI Images of the Pituitary Gland for Medical Image Analysis
CSI_7_PRO: DISSERTATION
Overview
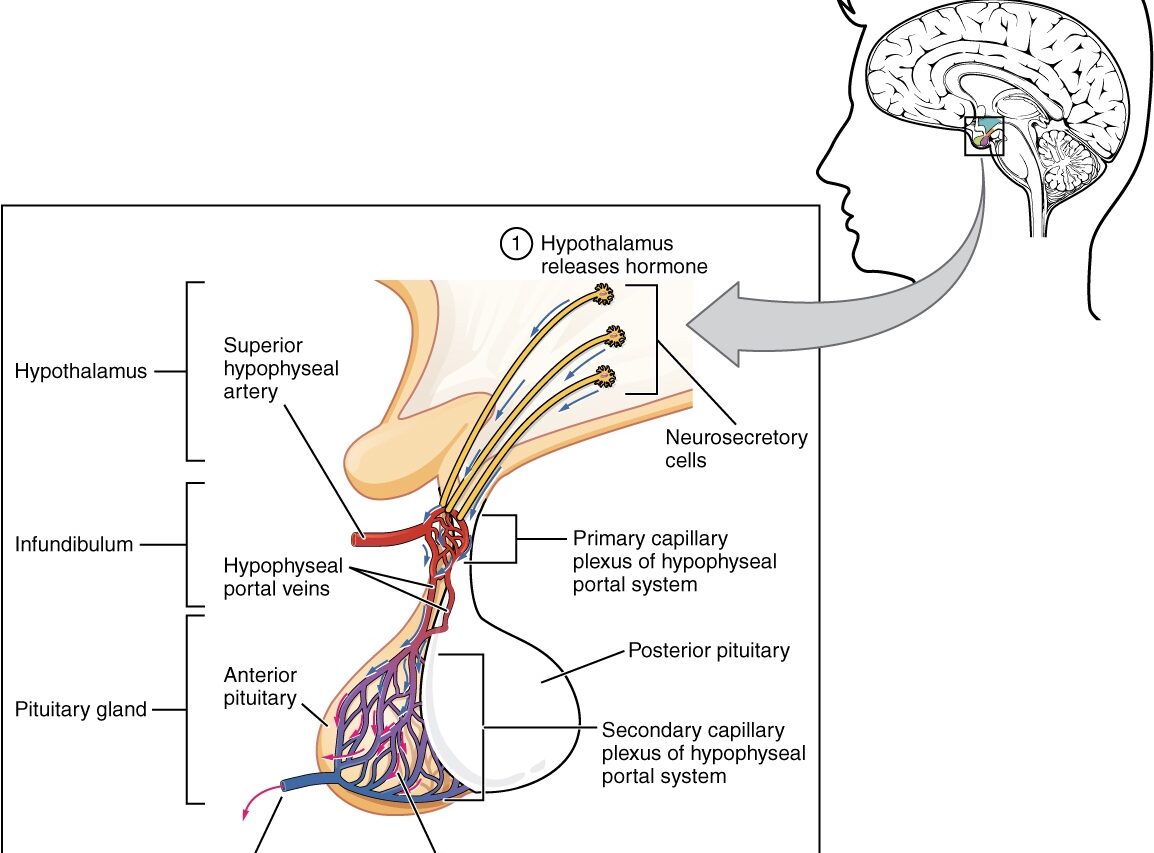
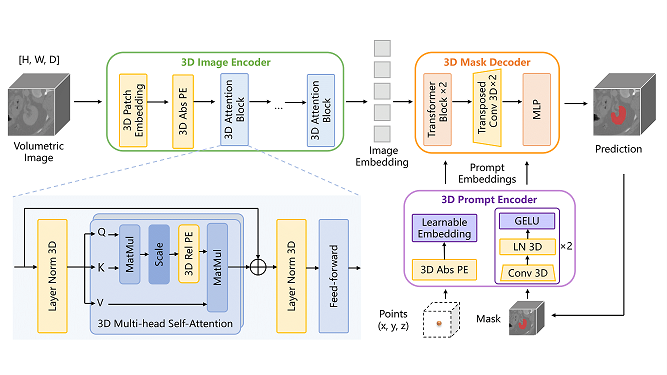
Imagine if doctors had a tool that could automatically identify and highlight the tiny pituitary gland in an MRI scan, making diagnoses faster and more reliable. That’s exactly what the Automatic Segmentation of 3D MRI Images of the Pituitary Gland project aimed to achieve using deep learning! The pituitary gland, also known as the “master gland,” controls many important hormones, but its small size and complex location make it challenging to identify in MRI scans. This project used SAM-Med3D, a pre-trained deep learning model based on the powerful nnU-Net architecture, to automatically segment the pituitary gland in 3D MRI brain scans. Along the way, we uncovered both the potential and the challenges of using AI in specialized medical image analysis.
Why Is This Important?
The pituitary gland plays a crucial role in regulating hormones that control everything from growth to metabolism. Accurate segmentation of this gland is key for diagnosing conditions like pituitary tumors. Manual segmentation, however, is time-consuming, tedious, and prone to human error. Traditional methods like thresholding or region growing often struggle due to the gland’s small size and the presence of noise in MRI scans. This project explored whether deep learning could take on this difficult task and make it more efficient and reliable for doctors.
How Did We Do It?
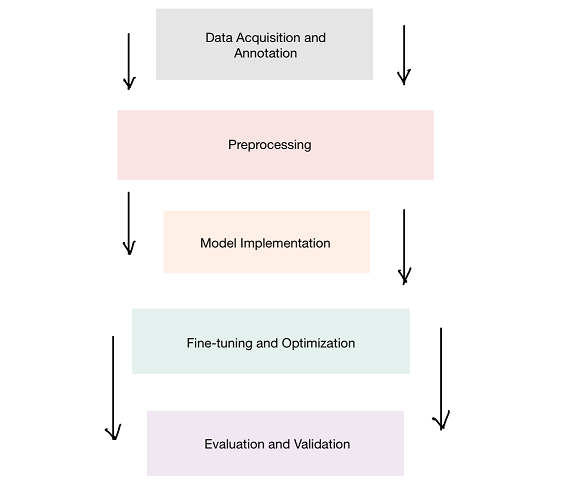
To address this problem, we worked with SAM-Med3D, a deep learning framework designed for medical imaging, and fine-tuned it to specifically segment the pituitary gland. Here’s how we approached it:
- Data Acquisition and Pre-processing
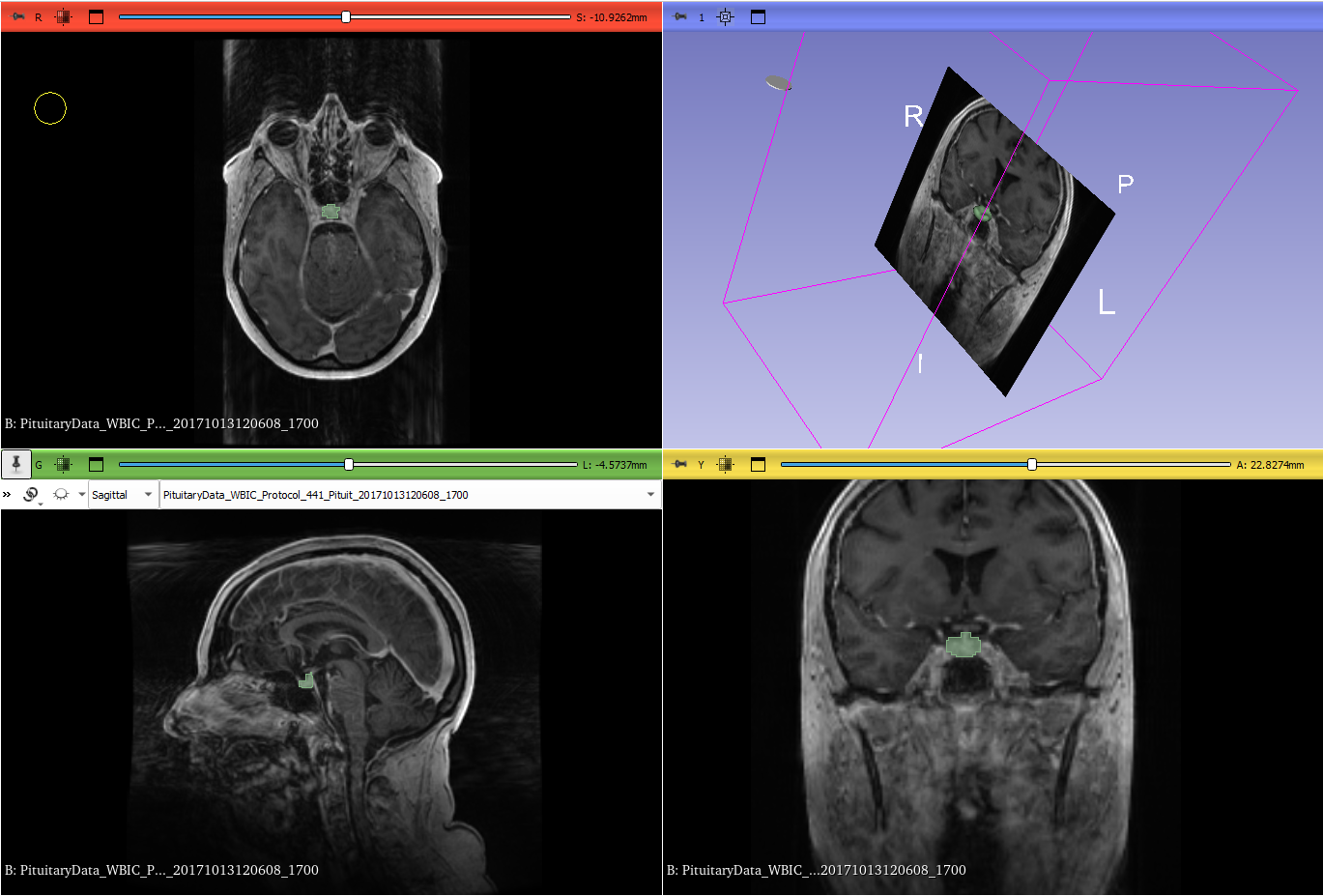
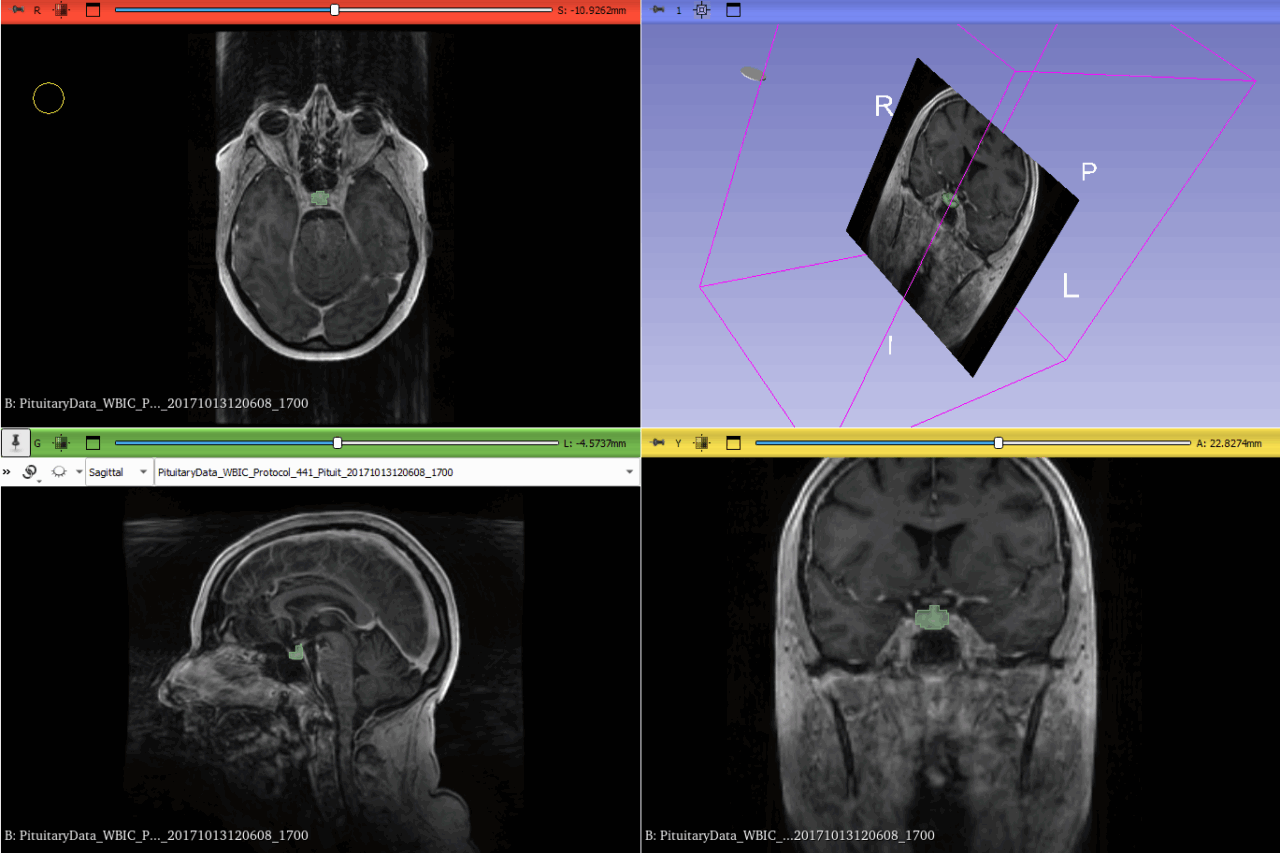
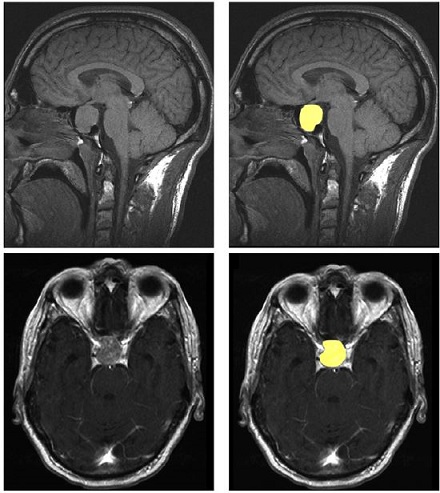
- Dataset: We used a collection of 3D MRI scans that included images of the pituitary gland. To create a benchmark, a subset of these images was manually segmented by expert radiologists. These served as the ground truth for training and testing the model.
- Normalization and Resampling: MRI scans often vary in intensity and size, making it crucial to normalize them. We used the Z-score method to ensure intensity values were consistent across all images, and resampled them to a uniform voxel size to make it easier for the model to learn effectively.
- Manual Ground Truth Creation: Expert radiologists manually labeled a portion of the scans, creating ground truth masks that were essential for training the model. This allowed us to evaluate how well the AI’s predictions matched expert judgment.
- Model Implementation
- Model Selection: We chose the SAM-Med3D model because it was built specifically for handling complex 3D medical images. Its nnU-Net architecture is well-known for its success in medical image segmentation.
- Transfer Learning: To make SAM-Med3D work for our specific needs, we used transfer learning. This means we took a model that was pre-trained on a large dataset and retrained just the final layers on our pituitary dataset. This allows the model to take advantage of the general image features it has already learned while focusing on the specifics of the pituitary gland.
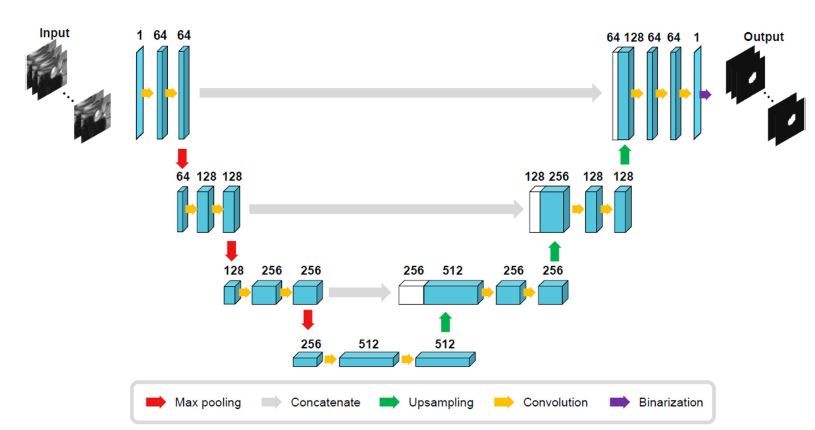
- Model Architecture: The architecture used was an encoder-decoder type with skip connections. These skip connections are crucial because they help maintain the spatial detail, which is particularly important for identifying small anatomical features like the pituitary gland.
- Training and Optimization
- Supervised Training: We trained the model using supervised learning, where the AI’s predictions were compared to the manual ground truth. To measure how well the model was learning, we used the Dice Loss function, which is great for segmentation tasks. To ensure our model could handle new data well, we used 5-fold cross-validation.
- Hyperparameter Tuning: Training an effective model isn’t just about the dataset—it’s also about the right parameters. We optimized things like learning rates, batch sizes, and number of epochs. We also used techniques like early stopping (to avoid overfitting by stopping training when it was no longer improving) and dropout (to prevent overfitting by randomly ignoring some neurons during training).
- Evaluation Metrics
To determine how well our model performed, we used several metrics:- Dice Similarity Coefficient (DSC): This is like a score for how well two images match each other. In our case, it measures the overlap between the AI’s segmented region and the actual pituitary gland segmented by experts. The formula is:
Where is the ground truth segmentation and is the model’s predicted segmentation.
- Precision and Recall: These metrics helped us understand how well the model was at finding the pituitary gland without falsely labeling other parts of the brain:
Where is true positives, is false positives, and is false negatives.
- Hausdorff Distance: This metric measures how far off the model’s boundaries are from the ground truth boundaries. It’s particularly useful for understanding how well the model handles complex shapes.
Results
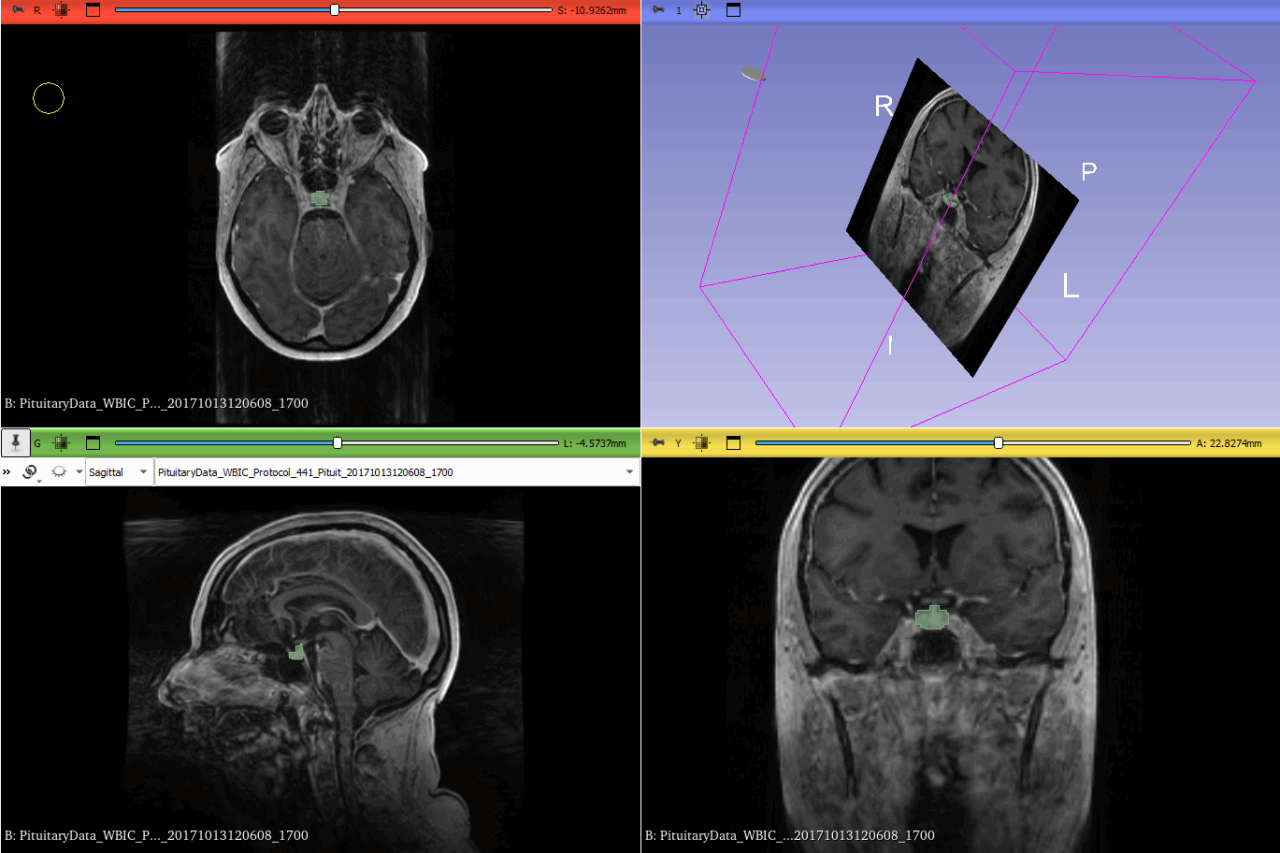
Even though our approach had promise, the SAM-Med3D model faced challenges in effectively segmenting the pituitary gland. The Dice Similarity Coefficient varied a lot, which meant the model wasn’t always consistent. This is likely due to the complexity of the gland’s shape and the differences between MRI scans of different patients. We achieved an average Dice score of 0.75, which, while decent, wasn’t quite up to the standards required for clinical reliability.
- Performance Evaluation: The model sometimes struggled, especially in cases where the gland had an unusual shape or where the MRI quality wasn’t optimal due to artifacts or patient movement. This highlights the difficulty of applying a general pre-trained model to a very specialized task without significant customization.
What Did We Learn?
- Model Limitations: The pre-trained SAM-Med3D model wasn’t perfectly suited for the pituitary gland without major adjustments. It became clear that for this kind of highly specialized segmentation, starting with a bespoke model or using advanced label preprocessing techniques might be necessary.
- Valuable Insights for Future Research: This project taught us that while pre-trained models are great starting points, they aren’t always the best fit for highly specialized medical applications. The importance of domain-specific training is evident.
Conclusion
This project highlighted both the potential and the challenges of using deep learning to segment the pituitary gland from MRI scans. While we made significant progress, the Automatic Segmentation of 3D MRI Images of the Pituitary Gland showed that more work is needed to reach clinical-grade accuracy. The results suggest that specialized training and further customization are crucial to achieving the precision required for clinical use.
Future Work
- Training from Scratch: Moving forward, we plan to train a model from scratch using a carefully curated dataset that specifically targets the pituitary gland. This would allow the model to learn more relevant features right from the start.
- Exploring Multi-modal Data: We also aim to incorporate additional imaging modalities, like CT scans, which could provide complementary information, making segmentation more robust.
- Addressing Computational Challenges: We need to focus on optimizing computational efficiency so that these models can be more easily used in clinical environments without the need for powerful hardware.
This thesis was supervised by Dr. Pawel Mackiewicz